TURBOMOLE#
TURBOMOLE is a commercial program package for ab initio electronic structure calculations. Unlike many other programs, the main focus in the development of TURBOMOLE has not been to implement all new methods and functionals, but to provide a fast and stable code which is able to treat molecules of industrial relevance at reasonable time and memory requirements.
TURBOMOLE has a frontend TmoleX that can work as a stand-alone or can be used to work remotely on the cluster.
NHR@FAU holds a binary site-license license for non-commercial academic use for TURBOMOLE 7.6 (and three update releases).
Availability / Target HPC systems#
- throughput cluster Woody: module
turbomoleandturbomole-parallel - parallel computer Meggie & Fritz:
turbomole/7.6(serial version) andturbomole/7.6-mpi(parallel version) - Use
module avail turbomoleto find the name of the TURBOMOLE module.
New versions of TURBOMOLE are installed by NHR@FAU upon request.
Notes#
TURBOMOLE with its fine grain IO can easily kill $FASTTMP. NHR@FAU
will kill / throttle any jobs which abuse the (parallel) file system.
Setup TmoleX for remote work on Woody-NG#
- Configurable under Extras -> Remote Systems
- Check the box Use queuing system and add the usual settings for
SBATCHin the field Script before job execution. Use--cpus-per-taskto adjust the number of cores for the job.
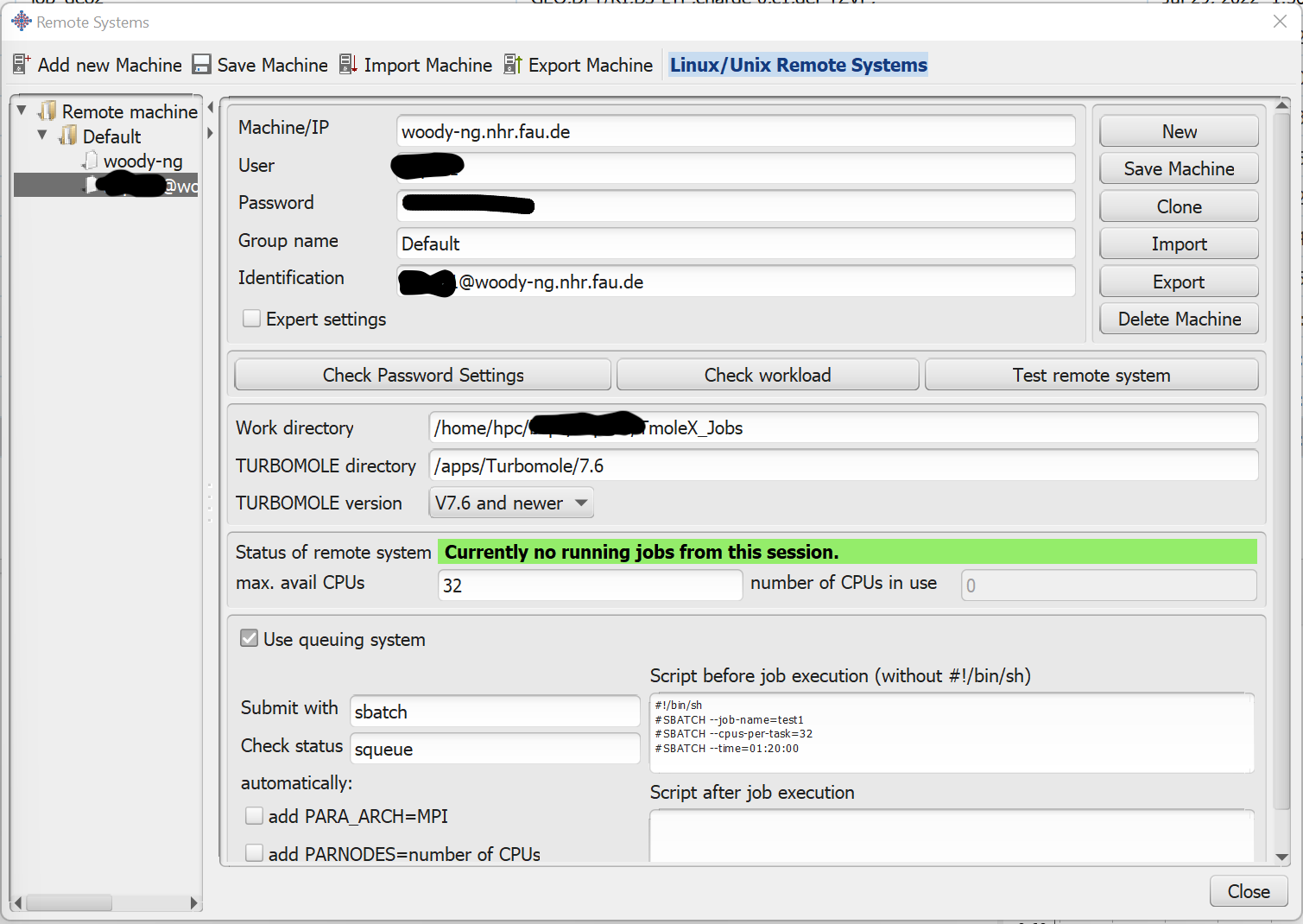
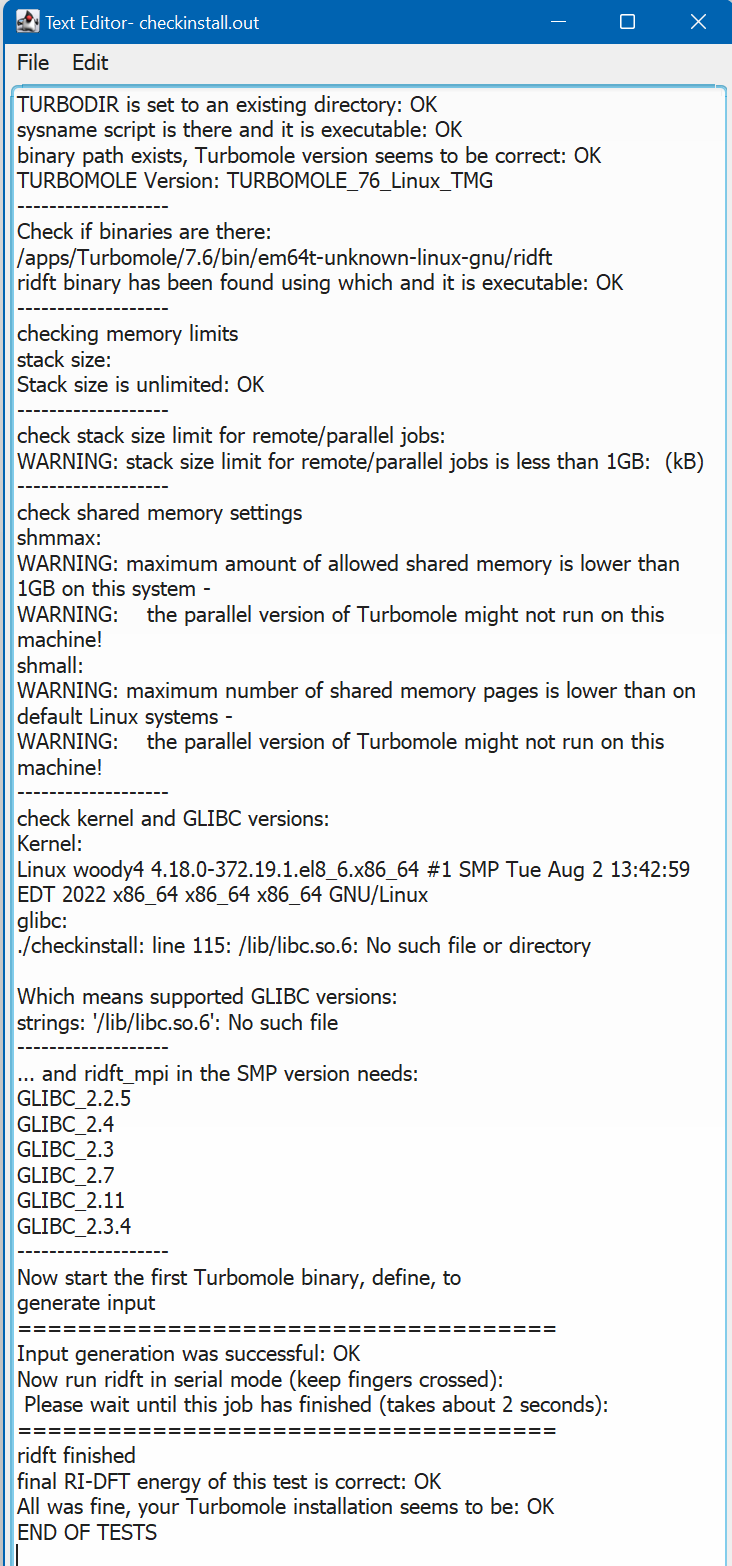
- Test the remote connection with the button Test remote system, to check that paths and parameters are correctly set.
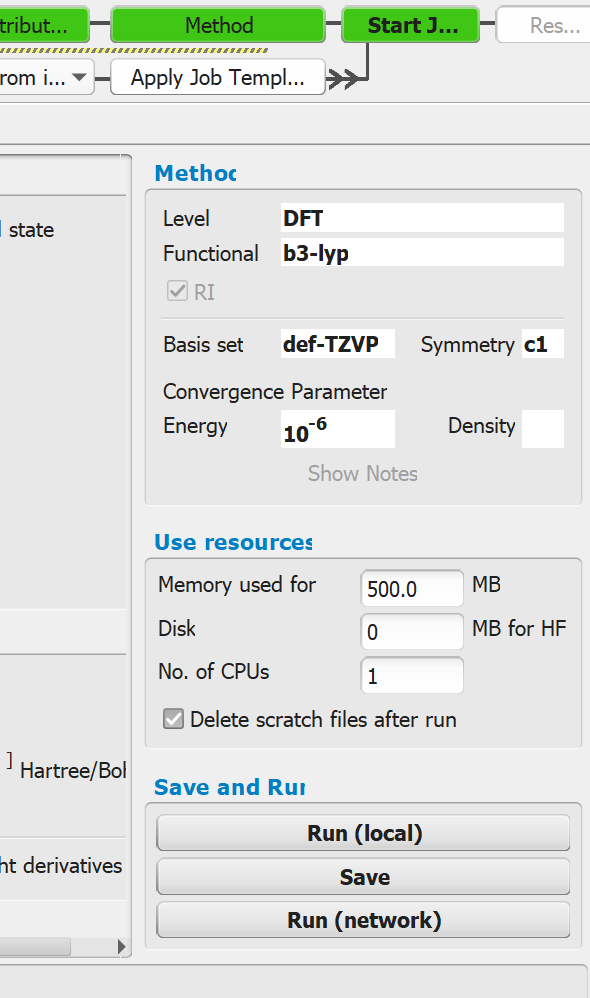
- Setup the job and start it via run (network).
- The queue status will pop up in another window.
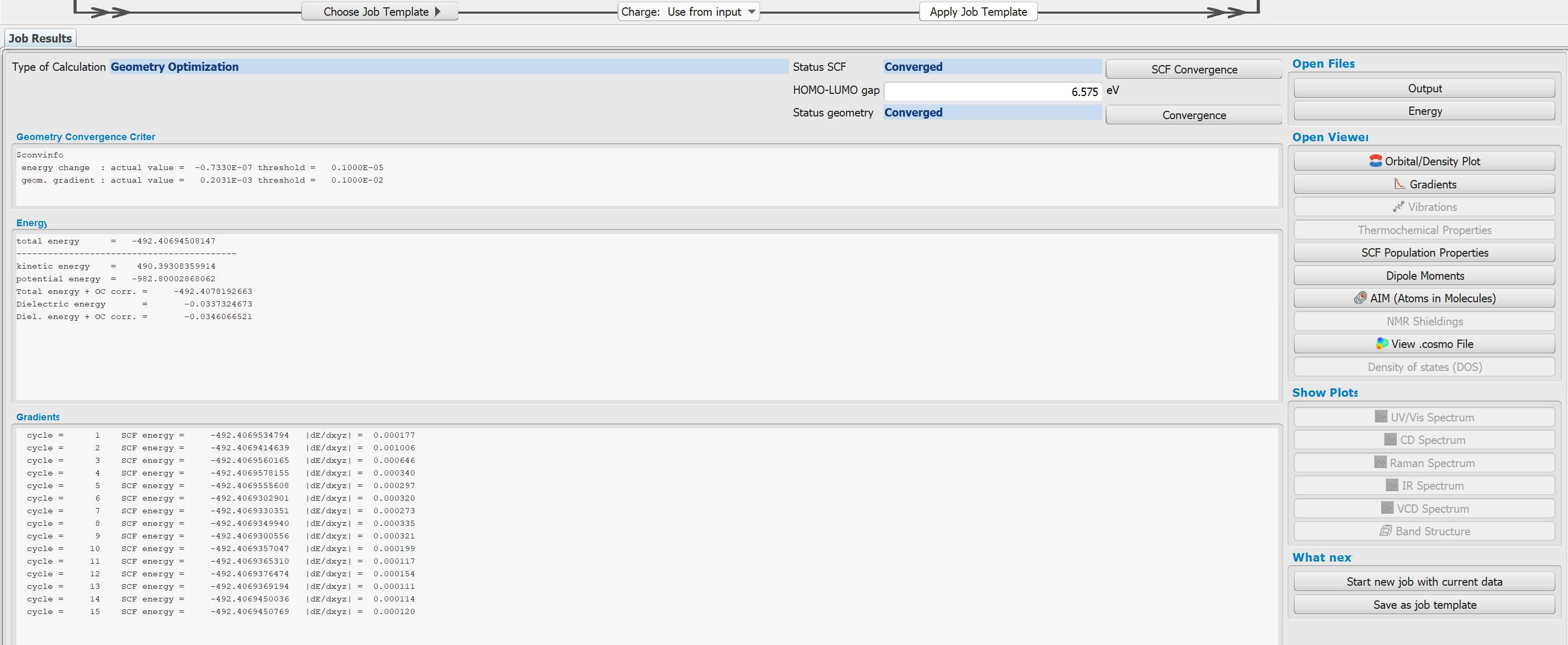
- When the job has finished, all data will be transferred to your local computer, the files will be deleted on the remote system and a short overview of the results will show up.